Performance of simuPOP in SeriationCT Across Platforms
| Created: 27 Jul 2015 | Modified: 30 Jun 2016 | BibTeX Entry | RIS Citation |
Background
SeriationCT uses the simuPOP framework, building custom actions on top of it but leveraging the excellent support Bo Peng has created for transmission modeling. simuPOP has support for running across multiple threads or cores, but this support has been disabled lately on Mac OS X given problems with OpenMP and GCC 4.2.1 (according to Bo).
Along with Bo, I’ve noted that OS X has excellent performance even without it, exceeding performance on Linux by a reasonable factor (sometimes even 2x).
Now that I’m running on my new simulation system, a 10-core Xeon E5-2650 V3 system equipped with a Xeon Phi 5110P coprocessor, I wanted to see what level of thread use in simuPOP provided the best performance.
The test setup runs the following simulation as the only active code on the system, apart from the MongoDB database which is serving as data repository. I ran this exact simulation with the same seed across thread count from 1 to 20, and then repeated this loop several times with different seeds, to generate multiple timing values for each thread count. The whole process was duplicated for 5 replicates and 10 replicates.
[mark@ewens sctcal-1]$ sim-seriationct-networkmodel.py --dbhost localhost --dbport 27017 --experiment sctcal-1 --popsize 250 --maxinittraits 5 --numloci 3 --innovrate 8.86017736123e-05 --simlength 12000 --debug 0 --seed 166602270 --reps 1 --samplefraction 0.5 --migrationfraction 0.0517857031302 --devel 0 --networkmodel networks/two-node-three-slice.zip --cores 10Single Replication
The results are shown below, with a loess smoothed trend line for each half of the graph. On the left, thread count is less than the number of physical cores, and it appears that threads are distributed among cores with scatter affinity (confirmed by observation in htop). On the right, hyperthreading is used to assign up to 2 threads per physical core, which is within the design spec of the E5-2650V3 processor. Some quick tests using 25 and 30 threads show that execution time shoots up precipitously given that execution is largely CPU-bound, and those data points are excluded in order to focus on the fine detail.
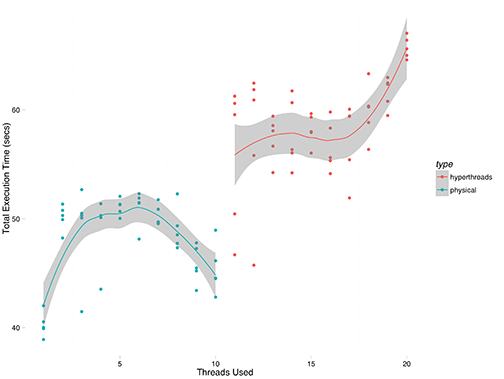
Immediately apparent is that the process really is CPU-bound, and that the code performs best with no hyperthreading, and a single thread per core. The strong increase around 18-20 threads is indicative of contention between the simulation threads and the MongoDB database which serves as the data repository, it looks like.
On the left, it is interesting that the absolute best performance is obtained by configuring simuPOP with a single thread, even with 10 physical cores present. Performance immediately worsens with 2 or more threads, before improving slightly between 8 and 10 threads. I have no explanation for why performance should improve, but clearly a single replication runs fastest when given a single execution thread, and the attempt to parallelize a single replication actually worsens the situation. I need to look at the simuPOP source and see why this occurs.
Performance with Multiple Replicates
I also ran tests with the same simulation command line, but with 5 replicates. Sometimes I use replicates with simuPOP, usually when I have a fixed grid of parameters and want to understand variability given the parameter space. Replicates are independent copies of a population, each evolving separately according to the same evolution schedule and operators, so this would seem to be a logical point at which parallelism would be leveraged.
This experiment I ran only a single series, given the longer execution times. The results are shown in Figure 2.
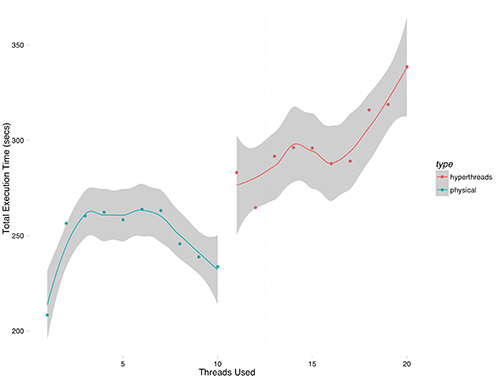
The pattern is the same, and the execution times are basically 5x the single replication time.
Parallelism in simuPOP
Looking at the version 1.1.6 source code, it appears that multiple threads are used in very specific situations. The simulator and its evolve function are single-threaded, with no OpenMP sections. Instead, parallelism is employed in specific loops, such as initializing a population or processing the individuals in a subpopulation, as in this example:
for (; sp != sp_end; ++sp) {
pop.activateVirtualSubPop(*sp);
size_t numValues = m_values.size();
if (numThreads() > 1 && !values.empty()) {
#ifdef _OPENMP
# pragma omp parallel firstprivate (idx)
{
size_t id = omp_get_thread_num();
IndIterator ind = pop.indIterator(sp->subPop(), id);
idx = idx + id * (pop.subPopSize(sp->subPop()) / numThreads());
for (; ind.valid(); ++ind, ++idx)
for (size_t i = 0; i < infoIdx.size(); ++i) {
ind->setInfo(values[idx % numValues], infoIdx[i]);
}
}
idx = idx + pop.subPopSize(sp->subPop());
#endifwhere tiles of individuals are processed by each of the N threads configured.
Examining the simulator itself, I can see why parallelism isn’t expressed at the replicate or subpopulation level. Applications built in simuPOP are capable of using pure-Python operators mixed with built-in C++ operations, and if replicates are threaded rather than being distributed across Unix processes, we run immediately into the GIL issue. So Bo has used OpenMP to parallelize things only within a single operator or function, which makes it safe.
What does this mean?
I’m probably not getting any speedup because the population size in my current simulation is small – 250 individuals – rather than the thousands to millions that one typically sees in genetics work. This means that the OpenMP and thread management overhead is dominating, and the more threads I use, the worse this gets. So, with small population sizes, the guideline here is always use a single thread.
Since simuPOP never parallelizes over replicates, it’s better to keep single replicates in their own Unix processes and use Grid Engine or a job scheduler to parallelize a set of simulations across the available cores.
The latter is what I’ve been doing on StarCluster in previous projects, so it’s a familiar and easily scriptable protocol. And, if I work with large population sizes at some point, the native threading in simuPOP could pay off, but otherwise, it’s best ignored for most of the work I’m doing.
